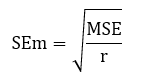The weblog is about estimation of genetic parameters like genotypic variance, phenotypic variance, heritability, genetic advance, genetic advance as a share of imply, phenotypic coefficient of variation (PCV), genotypic coefficient of variation (GCV) for the RCBD trails of genotypes. (Studying time 20 minutes).
1.
INTRODUCTION
In a normal
statistical context, a parameter
refers to a numerical attribute or attribute that describes a inhabitants.
It may be a hard and fast worth or an unknown amount that helps to explain or
summarize a particular side of a inhabitants. Genetic Parameter
is a statistical measure that quantifies the genetic contributions to traits
inside a inhabitants of an organism. Genetic parameter estimation in plant
breeding entails quantifying varied genetic elements that affect traits
of curiosity, comparable to yield, illness resistance or high quality attributes. These
parameters present important insights into the genetic foundation of those traits,
informing breeding selections geared toward bettering crop varieties.
Genetic parameters embody a
vary of measurements, together with heritability, genetic variance and genetic
advance. Heritability signifies the proportion of phenotypic
variation in a trait that’s attributable to genetic components, guiding breeders
on the potential response to choice. Genetic variance
quantifies the variability in traits because of genetic variations amongst
people, essential for understanding trait inheritance patterns. Genetic
advance measures the anticipated enchancment from choice, facilitating
environment friendly breeding methods. Understanding these genetic parameters empowers
plant breeders to develop improved cultivars tailor-made to particular agricultural
wants, enhancing crop productiveness, resilience and high quality. These parameters
are estimated via statistical analyses of trait knowledge collected from
breeding experiments, using methodologies comparable to variance element
evaluation and heritability estimation. The experiments are laid in varied experimental
designs that ensures legitimate and interpretable outcomes via randomization,
replication and management. Designs vary from easy fully randomized
designs to advanced ones like randomized full block designs (RCBD), factorial designs and Latin
squares. These designs assist isolate variable results and perceive their
interactions.
1. RANDOMIZED
COMPLETE BLOCK DESIGN
Randomized
Full Block Design (RCBD) is a basic experimental design used
extensively in plant breeding analysis to regulate for variability inside
experimental models. In RCBD, every block accommodates all genotypes, with random
project inside blocks, controlling for variability and making certain
complete genotype comparability. Therefore, it’s referred to as “Randomized
Full Block Design.” This design reduces experimental error and enhances
the precision of genotype imply comparisons by accounting for block-to-block
variability. It’s important for drawing legitimate inferences about genotype
results whereas minimizing the affect of extraneous components.
2.1 When
RCBD is used?
The
RCBD is employed in agricultural analysis underneath particular situations to realize
dependable and exact outcomes. Listed here are eventualities when RCBD is used: heterogeneous experimental models, identified gradients, a number of genotypes, restricted experimental models, small-scale
trials and so forth.
2.2 Assumptions
of RCBD
The
RCBD operates underneath a number of key assumptions to make sure legitimate and dependable
outcomes: homogeneity inside blocks,
independence of observations, additivity of results, random project,
normality, equal variance, no lacking knowledge and so forth.
2.3 Randomization steps in RCBD
Randomization
in a Randomized Full Block Design (RCBD) is a vital step to make sure
unbiased allocation of therapies to experimental models inside every block. Right here
are the detailed steps for randomization in RCBD:
- 1.
Determine the Remedies - 2.
Outline the Blocks - 3.
Assign Remedies Randomly inside Every Block - 4.
Document the Project - 5.
Repeat for All Blocks - 6.
Confirm Randomization - 7.
Create a Structure Plan
2.4 Evaluation
of Variance (ANOVA) for RCBD
In a RCRD, the Evaluation of Variance
(ANOVA) mannequin offers a
comparability by partitioning of variance because of varied sources.
It’s used to research the info and check the importance of genotype results. The statistical mannequin for ANOVA in RCBD is as
underneath:
Right here the null speculation is about as all genotypes means are equal
and the choice speculation is at the least one genotype pair differs
considerably. Significance of the imply sum of squares because of replications (Mr)
and genotypes (Mg) is examined towards error imply squares (Me). A comparability of
the calculated F (Mg/Me) with the important worth of F similar to genotype
levels of freedom and error levels of freedom provides the thought to just accept or
reject the null speculation.
2.5 Completely different statistic associated to RBD design
2.5.1 Commonplace error of imply (SEm):
2.5.2 Coefficient of Variation (CV%):
2.5.3 Important distinction at 5% stage of
significance
2.6 What if replication supply of
variation discovered vital in RCBD?
2.6.1 Causes for Important Replication in Plant Genotype Experiments
This contains environmental micro-variation (soil heterogeneity, microclimatic situations, and so forth.,), administration and cultural practices (inconsistent utility of therapies, variations in planting depth and spacing and so forth.,), biotic components (pest and illness strain, microbial exercise and so forth.,), phenotypic plasticity (adaptive responses), measurement and sampling error (human error in measurement, instrument calibration and so forth.,)
2.6.2 Addressing Important Replication in Plant Genotype Experiments
This may be achieved by bettering experimental design (improve block homogeneity, improve variety of replicates and so forth.,), standardize cultural practices (constant therapy utility, uniform planting strategies and so forth.,), management environmental components (monitor and handle microclimate, soil administration and so forth.,), common monitoring for biotic components (pest and illness administration, microbial inoculants and so forth.,), refine measurement strategies (coaching and calibration, automated measurements and so forth.,)
3 CALCULATION OF SIMPLE MEASURES OF VARIABILITY
Easy measures of
variability embrace vary, normal deviation, variance, normal deviation and
coefficient of variation. These measures assist in understanding the distribution
and unfold of knowledge, that are important for statistical evaluation and
decoding the variability inside an information set for given character.
3.1 Vary: The distinction
between the utmost and minimal values in an information set. Offers a fast sense of
the unfold of the info, however is delicate to outliers.
Vary = Most Worth – Minimal Worth
3.2 Commonplace Deviation (SD): A
measure of the typical distance of every knowledge level from the imply. Signifies how
unfold out the info factors are across the imply. A smaller SD signifies knowledge
factors are near the imply, whereas a bigger SD signifies they’re extra unfold
out.
The place, xi is every knowledge level, x ̅ is the imply of the info and n is the variety of knowledge factors
3.3 Variance: The
common of the squared variations from the imply. It measures the dispersion
of knowledge factors. It is the sq. of the usual deviation.
3.4 Coefficient of Variation (CV): The
ratio of the usual deviation to the imply, expressed as a share. It standardizes
the measure of variability by evaluating the usual deviation relative to the
imply. Helpful for evaluating the diploma of variation between completely different knowledge units,
particularly these with completely different models or broadly completely different means.
4. Variance
Parts
Within the context of plant breeding and genetics, ANOVA (Evaluation of
Variance) is commonly used to partition the noticed variance into completely different
elements: phenotypic variance, genotypic variance, and environmental
variance. These elements are essential for understanding the underlying
variability and for estimating the respective coefficients of variation.
4.4 What if genotypic variance is detrimental?
If σ2g
(genotypic variance) is detrimental, it signifies that the calculated worth just isn’t
possible since variance, by definition, can’t be detrimental. This example
sometimes arises because of small pattern dimension, massive experimental error, incorrect
knowledge or calculation and so forth. To deal with this points
improve replications, enhance experimental design, re-evaluate
knowledge and so forth. In abstract, a detrimental genotypic variance suggests the
want for a reassessment of the experimental design, knowledge high quality and evaluation
strategies.
5.
COEFFICIENTS OF VARIATION
5.1 Phenotypic Coefficient of Variation (PCV): Measures the extent of phenotypic variability relative to the imply of the trait.
5.2 Genotypic Coefficient of Variation (GCV): Measures the extent of genotypic variability relative to the imply of the trait.
5.3 Tips on how to Interpret the Relative
Values of GCV, PCV and ECV?
The relative values of
Genotypic Coefficient of Variation (GCV), Phenotypic Coefficient of Variation
(PCV), and Environmental Coefficient of Variation (ECV) present insights into
the sources and magnitude of variability inside a genetic inhabitants.
- GCV is Excessive In comparison with PCV: PCV sometimes exceeds or equals GCV because it
contains each genetic and environmental variance. If GCV surpasses PCV,
this implies a calculation error; evaluate for accuracy. - PCV is Excessive In comparison with GCV: PCV is larger than GCV, indicating
substantial environmental affect on the trait. The distinction suggests
vital environmental variance. Regardless of genetic variability, breeders
should reduce environmental results to pick successfully based mostly on genetic
potential. - ECV is Increased than GCV: The trait is closely influenced by
environmental components, with minimal genetic variability. Phenotypic
choice could also be tough. Introducing new genetic materials may assist
improve genetic variability and enhance choice effectivity for the
trait.
5.4 Tips on how to
Interpret Mixture of Values of GCV and PCV
- Excessive GCV and Excessive PCV: This
signifies that the trait is strongly influenced by genetic components, however
environmental components additionally play a big position. Regardless of the
environmental affect, the excessive genetic variability suggests good
potential for enchancment via choice. Deal with stabilizing the
setting to harness the genetic potential successfully. Breeders can
make vital progress by deciding on superior genotypes. - Excessive GCV and Low PCV: This
means that the trait is predominantly influenced by genetic components,
with minimal environmental influence. The excessive genetic variability just isn’t
masked by environmental results. This is a perfect scenario for breeders.
Choice can be extremely efficient for the reason that phenotypic efficiency
instantly displays the genetic potential. - Low GCV and Excessive PCV: This
signifies that the trait is basically influenced by environmental components,
with little genetic variability. The excessive phenotypic variability is usually
because of environmental results. Choice could be much less efficient as a result of
low genetic variability. Breeders could have to concentrate on bettering
environmental situations or administration practices to cut back the
environmental variance. Moreover, exploring wider genetic bases or
introducing new germplasm may very well be thought-about to extend genetic
variability. - Low GCV and Low PCV: This
means that the trait is comparatively secure with minimal affect from
each genetic and environmental components. The shortage of variability may
point out that the trait is both extremely conserved or has reached a
choice plateau. Restricted scope for enchancment via choice.
Breeders may have to introduce new genetic materials to extend
variability. Alternatively, focus may shift to different traits with larger
variability and potential for enchancment.
6.
Heritability and Genetic advance
Heritability and Genetic advance are vital
choice parameters. Heritability estimates together with the genetic advance are
usually extra useful in predicting genetic achieve underneath choice than
heritability estimates alone. Nonetheless, it isn’t needed {that a} character
exhibiting excessive heritability will even exhibit excessive genetic advance.
6.2 Tips on how to interpret the results of heritability in broad sense?
1. Low
Heritability (0-30%): A low share of phenotypic variation within the
trait is because of genetic components. A lot of the noticed variation is probably going due
to environmental influences. Selective breeding for this trait could be much less
efficient as a result of genetic variations contribute minimally to the trait’s
expression. As an alternative, concentrate on optimizing environmental situations to enhance
the trait.
2. Average Heritability (30-60%): A
average share of phenotypic variation is because of genetic components. Each
genetics and setting play vital roles in influencing the trait. Selective
breeding can result in average enhancements within the trait. Genetic good points may be
achieved, however additionally it is important to handle environmental components to totally
categorical the genetic potential.
Excessive Heritability (60% and above): A excessive
share of phenotypic variation is because of genetic components. A lot of the
variation within the trait may be attributed to genetic variations amongst
people. Selective breeding is extremely efficient for this trait. Important
genetic enhancements may be made, and the trait is much less influenced by
environmental components.
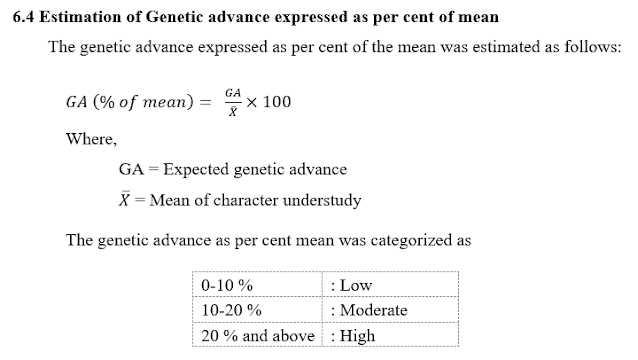
6.3 Estimation of Genetic advance (GA)
Genetic advance refers back to the enchancment in a trait achieved via choice. It is determined by the choice depth, heritability and phenotypic normal deviation of the trait. The anticipated genetic advance (GA) may be calculated for every character by adopting the next system at 5 % choice depth utilizing the fixed ‘Ok’ as 2.06.
6.5 Tips on how to Interpret the Results of Genetic Advance as Per Cent of Imply?
1. Low Genetic Advance (0-10%): The trait is much less aware of choice. Reaching vital genetic enchancment via choice alone could be difficult. It could be needed to think about different methods comparable to hybridization or bettering environmental situations.
2. Average Genetic Advance (10-20%): The trait reveals an inexpensive response to choice. Choice can result in noticeable enhancements within the trait. A balanced strategy of choice and environmental administration may be efficient.
3. Excessive Genetic Advance (20% and above): The trait is extremely aware of choice. Important genetic good points may be achieved via choice. This trait is a main candidate for intensive choice packages to realize speedy enchancment.
6.6 Combining The Outcomes of Heritability (Broad Sense) And Genetic Advance (As P.c of Imply)
Combining heritability (broad-sense heritability) and genetic advance as % of imply (GAM) offers a extra complete understanding of the potential for enchancment of traits in a breeding program. This mix helps in figuring out traits that aren’t solely genetically managed but in addition aware of choice.
SOLVED EXAMPLE
Dataset: The
experiment was laid in Randomized Full Block Design with three replications
in maize (Zea mays L.) by utilizing 30 genotypes. The info have been noticed
from every replication by randomly chosen vegetation for days to 50% flowering. Hyperlink of Dataset
|
Genotypes |
Replications |
Genotype whole |
Genotype imply |
||
|
R1 |
R2 |
R3 |
|||
|
G1 |
66 |
75 |
75 |
216.00 |
72.00 |
|
G2 |
68 |
75 |
76 |
219.00 |
73.00 |
|
G3 |
70 |
75 |
80 |
225.00 |
75.00 |
|
G4 |
70 |
81 |
86 |
237.00 |
79.00 |
|
G5 |
72 |
68 |
74 |
214.00 |
71.33 |
|
G6 |
66 |
72 |
80 |
218.00 |
72.67 |
|
G7 |
59 |
63 |
74 |
196.00 |
65.33 |
|
G8 |
66 |
69 |
79 |
214.00 |
71.33 |
|
G9 |
72 |
80 |
78 |
230.00 |
76.67 |
|
G10 |
64 |
66 |
83 |
213.00 |
71.00 |
|
G11 |
84 |
72 |
74 |
230.00 |
76.67 |
|
G12 |
60 |
64 |
75 |
199.00 |
66.33 |
|
G13 |
62 |
68 |
65 |
195.00 |
65.00 |
|
G14 |
63 |
72 |
75 |
210.00 |
70.00 |
|
G15 |
73 |
81 |
70 |
224.00 |
74.67 |
|
G16 |
58 |
84 |
70 |
212.00 |
70.67 |
|
G17 |
77 |
82 |
86 |
245.00 |
81.67 |
|
G18 |
64 |
69 |
75 |
208.00 |
69.33 |
|
G19 |
82 |
82 |
84 |
248.00 |
82.67 |
|
G20 |
72 |
74 |
75 |
221.00 |
73.67 |
|
G21 |
75 |
80 |
78 |
233.00 |
77.67 |
|
G22 |
70 |
76 |
82 |
228.00 |
76.00 |
|
G23 |
76 |
83 |
82 |
241.00 |
80.33 |
|
G24 |
77 |
76 |
75 |
228.00 |
76.00 |
|
G25 |
77 |
83 |
70 |
230.00 |
76.67 |
|
G26 |
76 |
84 |
86 |
246.00 |
82.00 |
|
G27 |
83 |
68 |
72 |
223.00 |
74.33 |
|
G28 |
61 |
75 |
84 |
220.00 |
73.33 |
|
G29 |
67 |
78 |
60 |
205.00 |
68.33 |
|
G30 |
67 |
70 |
78 |
215.00 |
71.67 |
|
Replication whole |
2097 |
2245 |
2301 |
|
|
|
Grand whole |
6643 |
||||
7.1 Evaluation of Variance
Null speculation for genotypes and replication
H0: There are not any vital variations amongst technique of genotypes underneath examine.
Ha: There are not any vital variations amongst technique of replications underneath examine.
Conclusion:
• Low GCV and low PCV for days to 50% flowering point out low variability. The shortage of variability may point out that the trait is both extremely conserved or has reached a variety plateau.
• Heritability is <30 indicated extra affect of setting within the inheritance of the trait
• Low heritability coupled with low genetic advance as per cent of imply point out the choice wouldn’t be rewarding because of environmental fluctuations
7.
STEPS TO
PERFORM ANALYSIS OF GENETIC PARAMETER ESTIMATION IN AGRI ANALYZE
Step 1: To create a CSV file with columns for Genotype, replication and trait
(DFF). Hyperlink of Dataset
Step 2: Go together with Agri Analyze website. https://agrianalyze.com/Default.aspx Register by utilizing e mail and cellular quantity
Step 3: Click on on
ANALYTICAL TOOL
Step 4: Click on on GENETICS
AND PLANT BREEDING
Step 5: Click on on GENETIC
PARAMETER ESTIMATION
Step 6: Add the CSV file and choose Genotypes, Replication and Click on on Submit
Gomez, Ok. A., & Gomez, A. A. (1984). Statistical Procedures for Agricultural Analysis. John wiley & sons. 25-30.
Singh, P. and Narayanan, S.S. (1993) Biometrical Methods in Plant Breeding. New Delhi, India: Kalyani Publishers.
Weblog Credit score: