Swap in one other LLM
There are a number of methods to run the identical activity with a special mannequin. First, create a brand new chat object with that totally different mannequin. Right here’s the code for testing Google Gemini 3 Flash Preview:
my_chat_gemini <- chat_google_gemini(mannequin = "gemini-3-flash-preview")Then you’ll be able to run the duty in one among 3 ways.
1. Clone an current activity and add the chat as its solver with $set_solver():
my_task_gemini <- my_task$clone()
my_task_gemini$set_solver(generate(my_chat_gemini))
my_task_gemini$eval(epochs = 3)2. Clone an current activity and add the brand new chat as a solver once you run it:
my_task_gemini <- my_task$clone()
my_task_gemini$eval(epochs = 3, solver_chat = my_chat_gemini)3. Create a brand new activity from scratch, which lets you embrace a brand new identify:
my_task_gemini <- Process$new(
dataset = my_dataset,
solver = generate(my_chat_gemini),
scorer = model_graded_qa(
partial_credit = FALSE,
scorer_chat = ellmer::chat_anthropic(mannequin = "claude-opus-4-6")
),
identify = "Gemini flash 3 preview"
)
my_task_gemini$eval(epochs = 3)Be sure you’ve set your API key for every supplier you need to check, except you’re utilizing a platform that doesn’t want them, comparable to native LLMs with ollama.
View a number of activity runs
When you’ve run a number of duties with totally different fashions, you need to use the vitals_bind() perform to mix the outcomes:
both_tasks <- vitals_bind(
gpt5_nano = my_task,
gemini_3_flash = my_task_gemini
)Instance of mixed activity outcomes working every LLM with three epochs.
Sharon Machlis
This returns an R information body with columns for activity, id, epoch, rating, and metadata. The metadata column comprises a knowledge body in every row with columns for enter, goal, outcome, solver_chat, scorer_chat, scorer_metadata, and scorer.
To flatten the enter, goal, and outcome columns and make them simpler to scan and analyze, I un-nested the metadata column with:
library(tidyr)
both_tasks_wide <- both_tasks |>
unnest_longer(metadata) |>
unnest_wider(metadata)I used to be then in a position to run a fast script to cycle by means of every bar-chart outcome code and see what it produced:
library(dplyr)
# Some outcomes are surrounded by markdown and that markdown code must be eliminated or the R code will not run
extract_code <- perform(textual content) ```n
# Filter for barchart outcomes solely
barchart_results <- both_tasks_wide |>
filter(id == "barchart")
# Loop by means of every outcome
for (i in seq_len(nrow(barchart_results))) {
code_to_run <- extract_code(barchart_results$outcome[i])
rating <- as.character(barchart_results$rating[i])
task_name <- barchart_results$activity[i]
epoch <- barchart_results$epoch[i]
# Show information
cat("n", strrep("=", 60), "n")
cat("Process:", task_name, "| Epoch:", epoch, "| Rating:", rating, "n")
cat(strrep("=", 60), "nn")
# Attempt to run the code and print the plot
tryCatch(
{
plot_obj <- eval(parse(textual content = code_to_run))
print(plot_obj)
Sys.sleep(3)
},
error = perform(e) {
cat("Error working code:", e$message, "n")
Sys.sleep(3)
}
)
}
cat("nFinished displaying all", nrow(barchart_results), "bar charts.n")Take a look at native LLMs
That is one among my favourite use instances for vitals. Presently, fashions that match into my PC’s 12GB of GPU RAM are fairly restricted. However I’m hopeful that small fashions will quickly be helpful for extra duties I’d love to do domestically with delicate information. Vitals makes it straightforward for me to check new LLMs on a few of my particular use instances.
vitals (through ellmer) helps ollama, a well-liked manner of working LLMs domestically. To make use of ollama, obtain, set up, and run the ollama software, and both use the desktop app or a terminal window to run it. The syntax is ollama pull to obtain an LLM, or ollama run to each obtain and begin a chat in the event you’d like to verify the mannequin works in your system. For instance: ollama pull ministral-3:14b.
The rollama R package deal enables you to obtain a neighborhood LLM for ollama inside R, so long as ollama is working. The syntax is rollama::pull_model("model-name"). For instance, rollama::pull_model("ministral-3:14b"). You’ll be able to check whether or not R can see ollama working in your system with rollama::ping_ollama().
I additionally pulled Google’s gemma3-12b and Microsoft’s phi4, then created duties for every of them with the identical dataset I used earlier than. Notice that as of this writing, you want the dev model of vitals to deal with LLM names that embrace colons (the subsequent CRAN model after 0.2.0 ought to deal with that, although):
# Create chat objects
ministral_chat <- chat_ollama(
mannequin = "ministral-3:14b"
)
gemma_chat <- chat_ollama(
mannequin = "gemma3:12b"
)
phi_chat <- chat_ollama(
mannequin = "phi4"
)
# Create one activity with ministral, with out naming it
ollama_task <- Process$new(
dataset = my_dataset,
solver = generate(ministral_chat),
scorer = model_graded_qa(
scorer_chat = ellmer::chat_anthropic(mannequin = "claude-opus-4-6")
)
)
# Run that activity object's evals
ollama_task$eval(epochs = 5)
# Clone that activity and run it with totally different LLM chat objects
gemma_task <- ollama_task$clone()
gemma_task$eval(epochs = 5, solver_chat = gemma_chat)
phi_task <- ollama_task$clone()
phi_task$eval(epochs = 5, solver_chat = phi_chat)
# Flip all these outcomes right into a mixed information body
ollama_tasks <- vitals_bind(
ministral = ollama_task,
gemma = gemma_task,
phi = phi_task
)All three native LLMs nailed the sentiment evaluation, and all did poorly on the bar chart. Some code produced bar charts however not with axes flipped and sorted in descending order; different code didn’t work in any respect.
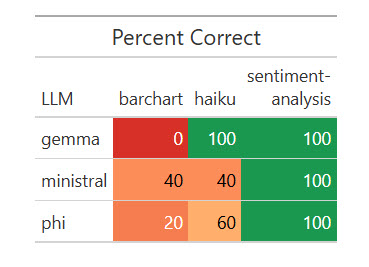
Outcomes of 1 run of my dataset with 5 native LLMs.
Sharon Machlis
R code for the outcomes desk above:
library(dplyr)
library(gt)
library(scales)
# Put together the info
plot_data <- ollama_tasks |>
rename(LLM = activity, activity = id) |>
group_by(LLM, activity) |>
summarize(
pct_correct = imply(rating == "C") * 100,
.teams = "drop"
)
color_fn <- col_numeric(
palette = c("#d73027", "#fc8d59", "#fc8d59", "#fee08b", "#1a9850"),
area = c(0, 20, 40, 60, 100)
)
plot_data |>
tidyr::pivot_wider(names_from = activity, values_from = pct_correct) |>
gt() |>
tab_header(title = "P.c Appropriate") |>
cols_label(`sentiment-analysis` = html("sentiment-
evaluation")) |>
data_color(
columns = -LLM,
fn = color_fn
)It value me 39 cents for Opus to guage these native LLM runs—not a foul discount.
Extract structured information from textual content
Vitals has a particular perform for extracting structured information from plain textual content: generate_structured(). It requires each a chat object and an outlined information kind you need the LLM to return. As of this writing, you want the event model of vitals to make use of the generate_structured() perform.
First, right here’s my new dataset to extract subject, speaker identify and affiliation, date, and begin time from a plain-text description. The extra complicated model asks the LLM to transform the time zone to Japanese Time from Central European Time:
extract_dataset <- information.body(
id = c("entity-extract-basic", "entity-extract-more-complex"),
enter = c(
"Extract the workshop subject, speaker identify, speaker affiliation, date in 'yyyy-mm-dd' format, and begin time in 'hh:mm' format from the textual content under. Assume the date yr makes essentially the most sense provided that at the moment's date is February 7, 2026. Return ONLY these entities within the format {subject}, {speaker identify}, {date}, {start_time}. R Bundle Growth in PositronrnThursday, January fifteenth, 18:00 - 20:00 CET (Rome, Berlin, Paris timezone) rnStephen D. Turner is an affiliate professor of information science on the College of Virginia Faculty of Information Science. Previous to re-joining UVA he was a knowledge scientist in nationwide safety and protection consulting, and later at a biotech firm (Colossal, the de-extinction firm) the place he constructed and deployed scores of R packages. ",
"Extract the workshop subject, speaker identify, speaker affiliation, date in 'yyyy-mm-dd' format, and begin time in Japanese Time zone in 'hh:mm ET' format from the textual content under. (TZ is the time zone). Assume the date yr makes essentially the most sense provided that at the moment's date is February 7, 2026. Return ONLY these entities within the format {subject}, {speaker identify}, {date}, {start_time}. Convert the given time to Japanese Time if required. R Bundle Growth in PositronrnThursday, January fifteenth, 18:00 - 20:00 CET (Rome, Berlin, Paris timezone) rnStephen D. Turner is an affiliate professor of information science on the College of Virginia Faculty of Information Science. Previous to re-joining UVA he was a knowledge scientist in nationwide safety and protection consulting, and later at a biotech firm (Colossal, the de-extinction firm) the place he constructed and deployed scores of R packages. "
),
goal = c(
"R Bundle Growth in Positron, Stephen D. Turner, College of Virginia (or College of Virginia Faculty of Information Science), 2026-01-15, 18:00. OR R Bundle Growth in Positron, Stephen D. Turner, College of Virginia (or College of Virginia Faculty of Information Science), 2026-01-15, 18:00 CET.",
"R Bundle Growth in Positron, Stephen D. Turner, College of Virginia (or College of Virginia Faculty of Information Science), 2026-01-15, 12:00 ET."
)
)Beneath is an instance of the right way to outline a knowledge construction utilizing ellmer’s type_object() perform. Every of the arguments provides the identify of a knowledge subject and its kind (string, integer, and so forth). I’m specifying I need to extract a workshop_topic, speaker_name, current_speaker_affiliation, date (as a string), and start_time (additionally as a string):
my_object <- type_object(
workshop_topic = type_string(),
speaker_name = type_string(),
current_speaker_affiliation = type_string(),
date = type_string(
"Date in yyyy-mm-dd format"
),
start_time = type_string(
"Begin time in hh:mm format, with timezone abbreviation if relevant"
)
)Subsequent, I’ll use the chat objects I created earlier in a brand new structured information activity, utilizing Sonnet because the choose since grading is simple:
my_task_structured <- Process$new(
dataset = extract_dataset,
solver = generate_structured(
solver_chat = my_chat,
kind = my_object
),
scorer = model_graded_qa(
partial_credit = FALSE,
scorer_chat = ellmer::chat_anthropic(mannequin = "claude-sonnet-4-6")
)
)
gemini_task_structured <- my_task_structured$clone()
# You'll want to add the sort to generate_structured(), that is not included when a structured activity is cloned
gemini_task_structured$set_solver(
generate_structured(solver_chat = my_chat_gemini, kind = my_object)
)
ministral_task_structured <- my_task_structured$clone()
ministral_task_structured$set_solver(
generate_structured(solver_chat = ministral_chat, kind = my_object)
)
phi_task_structured <- my_task_structured$clone()
phi_task_structured$set_solver(
generate_structured(solver_chat = phi_chat, kind = my_object)
)
gemma_task_structured <- my_task_structured$clone()
gemma_task_structured$set_solver(
generate_structured(
solver_chat = gemma_chat,
kind = my_object
)
)
# Run the evaluations!
my_task_structured$eval(epochs = 3)
gemini_task_structured$eval(epochs = 3)
ministral_task_structured$eval(epochs = 3)
gemma_task_structured$eval(epochs = 3)
phi_task_structured$eval(epochs = 3)
# Save outcomes to information body
structured_tasks <- vitals_bind(
gemini = gemini_task_structured,
gpt_5_nano = my_task_structured,
ministral = ministral_task_structured,
gemma = gemma_task_structured,
phi = phi_task_structured
)
saveRDS(structured_tasks, "structured_tasks.Rds")It value me 16 cents for Sonnet to guage 15 analysis runs of two queries and outcomes every.
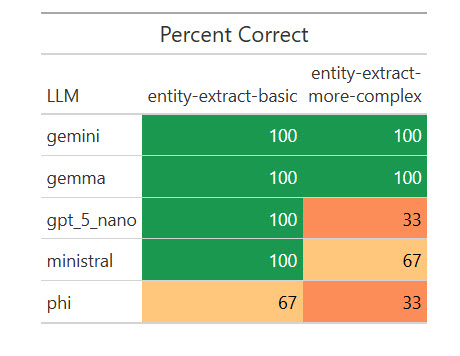
Listed below are the outcomes:
How varied LLMs fared on extracting structured information from textual content.
Sharon Machlis
I used to be shocked {that a} native mannequin, Gemma, scored 100%. I wished to see if that was a fluke, so I ran the eval one other 17 occasions for a complete of 20. Weirdly, it missed on two of the 20 primary extractions by giving the title as “R Bundle Growth” as a substitute of “R Bundle Growth in Positron,” however scored 100% on the extra complicated ones. I requested Claude Opus about that, and it mentioned my “simpler” activity was extra ambiguous for a much less succesful mannequin to grasp. Vital takeaway: Be as particular as potential in your directions!
Nonetheless, Gemma’s outcomes have been ok on this activity for me to think about testing it on some real-world entity extraction duties. And I wouldn’t have recognized that with out working automated evaluations on a number of native LLMs.
Conclusion
For those who’re used to writing code that offers predictable, repeatable responses, a script that generates totally different solutions every time it runs can really feel unsettling. Whereas there are not any ensures relating to predicting an LLM’s subsequent response, evals can improve your confidence in your code by letting you run structured checks with measurable responses, as a substitute of testing through handbook, ad-hoc queries. And, because the mannequin panorama retains evolving, you’ll be able to keep present by testing how newer LLMs carry out—not on generic benchmarks, however on the duties that matter most to you.

