Genomic prediction and design now require fashions that join native motifs with megabase scale regulatory context and that function throughout many organisms. Nucleotide Transformer v3, or NTv3, is InstaDeep’s new multi species genomics basis mannequin for this setting. It unifies illustration studying, useful monitor and genome annotation prediction, and controllable sequence era in a single spine that runs on 1 Mb contexts at single nucleotide decision.
Earlier Nucleotide Transformer fashions already confirmed that self supervised pretraining on 1000’s of genomes yields robust options for molecular phenotype prediction. The unique sequence included fashions from 50M to 2.5B parameters educated on 3,200 human genomes and 850 extra genomes from numerous species. NTv3 retains this sequence solely pretraining thought however extends it to longer contexts and provides express useful supervision and a generative mode.
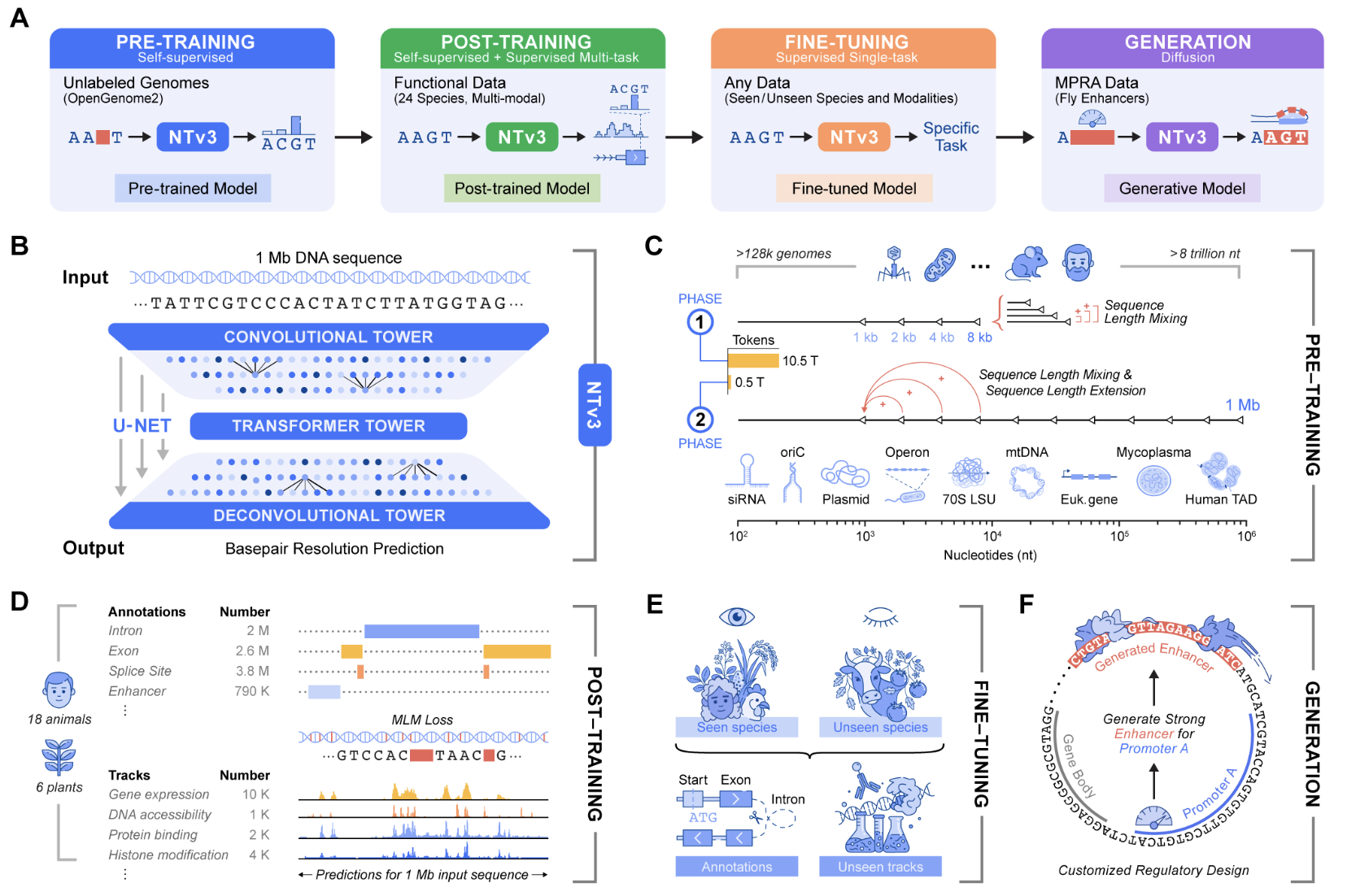
Structure for 1 Mb genomic home windows
NTv3 makes use of a U-Web type structure that targets very lengthy genomic home windows. A convolutional downsampling tower compresses the enter sequence, a transformer stack fashions lengthy vary dependencies in that compressed house, and a deconvolution tower restores base stage decision for prediction and era. Inputs are tokenized on the character stage over A, T, C, G, N with particular tokens equivalent to
The smallest public mannequin, NTv3 8M pre, has about 7.69M parameters with hidden dimension 256, FFN dimension 1,024, 2 transformer layers, 8 consideration heads, and seven downsample levels. On the excessive finish, NTv3 650M makes use of hidden dimension 1,536, FFN dimension 6,144, 12 transformer layers, 24 consideration heads, and seven downsample levels, and provides conditioning layers for species particular prediction heads.
Coaching information
The NTv3 mannequin is pretrained on 9 trillion base pairs from the OpenGenome2 useful resource utilizing base decision masked language modeling. After this stage, the mannequin is publish educated with a joint goal that integrates continued self supervision with supervised studying on roughly 16,000 useful tracks and annotation labels from 24 animal and plant species.
Efficiency and Ntv3 Benchmark
After publish coaching NTv3 achieves cutting-edge accuracy for useful monitor prediction and genome annotation throughout species. It outperforms robust sequence to operate fashions and former genomic basis fashions on current public benchmarks and on the brand new Ntv3 Benchmark, which is outlined as a managed downstream advantageous tuning suite with standardized 32 kb enter home windows and base decision outputs.
The Ntv3 Benchmark presently consists of 106 lengthy vary, single nucleotide, cross assay, cross species duties. As a result of NTv3 sees 1000’s of tracks throughout 24 species throughout publish coaching, the mannequin learns a shared regulatory grammar that transfers between organisms and assays and helps coherent lengthy vary genome to operate inference.
From prediction to controllable sequence era
Past prediction, NTv3 might be advantageous tuned right into a controllable generative mannequin through masked diffusion language modeling. On this mode the mannequin receives conditioning alerts that encode desired enhancer exercise ranges and promoter selectivity, and it fills masked spans within the DNA sequence in a manner that’s in line with these situations.
In experiments described within the launch supplies, the group designs 1,000 enhancer sequences with specified exercise and promoter specificity and validates them in vitro utilizing STARR seq assays in collaboration with the Stark Lab. The outcomes present that these generated enhancers recuperate the meant ordering of exercise ranges and attain greater than 2 occasions improved promoter specificity in contrast with baselines.
Comparability Desk
| Dimension | NTv3 (Nucleotide Transformer v3) | GENA-LM |
|---|---|---|
| Main purpose | Unified multi species genomics basis mannequin for illustration studying, sequence to operate prediction and controllable sequence era | Household of DNA language fashions for lengthy sequences centered on switch studying for a lot of supervised genomic prediction duties |
| Structure | U-Web type convolutional tower, transformer stack, deconvolutional tower, single base decision language mannequin, publish educated variations add multi species conditioning and process particular heads | BERT primarily based encoder fashions with 12 or 24 layers and BigBird variants with sparse consideration, prolonged additional with recurrent reminiscence transformer for lengthy contexts |
| Parameter scale | Household spans 8M, 100M and 650M parameters | Base fashions have 110M parameters and huge fashions have 336M parameters, together with BigBird variants at 110M |
| Native context size | As much as 1 Mb enter at single nucleotide decision for each pre educated and publish educated fashions | As much as about 4500 bp with 512 BPE tokens for BERT fashions and as much as 36000 bp with 4096 tokens for BigBird fashions |
| Prolonged context mechanism | Makes use of U-Web type convolutional tower to mixture lengthy vary context earlier than transformer layers whereas holding single base decision; context size is fastened at 1 Mb within the launched checkpoints | Makes use of sparse consideration in BigBird variants plus recurrent reminiscence transformer to increase efficient context to tons of of 1000’s of base pairs |
| Tokenization | Character stage tokenizer over A, T, C, G, N and particular tokens; every nucleotide is a token | BPE tokenizer on DNA that maps to about 4500 bp for 512 tokens; two tokenizers are used, one on T2T solely and one on T2T plus 1000G SNPs plus multispecies information |
| Pretraining corpus measurement | First stage pre coaching on OpenGenome2 with about 9 trillion base pairs from greater than 128000 species | Human solely fashions educated on pre processed human T2T v2 plus 1000 Genomes SNPs, about 480 × 10^9 base pairs, multispecies fashions educated on mixed human and multispecies information, about 1072 × 10^9 base pairs |
| Species protection | Greater than 128000 species in OpenGenome2 pretraining and publish coaching supervision from 24 animal and plant species | Human centered fashions plus taxon particular fashions for yeast, Arabidopsis and Drosophila and multispecies fashions from ENSEMBL genomes |
| Supervised publish coaching alerts | About 16000 useful tracks throughout about 10 assay sorts and about 2700 tissues in 24 species, used to situation the spine with discrete labels and to coach useful heads | Tremendous tuned on a number of supervised duties, together with promoters, splice websites, Drosophila enhancers, chromatin profiles and polyadenylation websites, with process particular heads on high of the LM |
| Generative capabilities | May be advantageous tuned right into a controllable generative mannequin utilizing masked diffusion language modeling, used to design 1000 promoter particular enhancers that achieved greater than 2× elevated specificity in STARR seq assays | Primarily used as a masked language mannequin and have extractor, helps sequence completion via MLM however the principle publication focuses on predictive duties fairly than express controllable sequence design |
Key Takeaways
- NTv3 is a protracted vary, multi species genomics basis mannequin: It unifies illustration studying, useful monitor prediction, genome annotation, and controllable sequence era in a single U Web type structure that helps 1 Mb nucleotide decision context throughout 24 animal and plant species.
- The mannequin is educated on 9 trillion base pairs with joint self supervised and supervised targets: NTv3 is pretrained on 9 trillion base pairs from OpenGenome2 with base decision masked language modeling, then publish educated on greater than 16,000 useful tracks and annotation labels from 24 species utilizing a joint goal that mixes continued self supervision with supervised studying.
- NTv3 achieves cutting-edge efficiency on the Ntv3 Benchmark: After publish coaching, NTv3 reaches cutting-edge accuracy for useful monitor prediction and genome annotation throughout species and outperforms earlier sequence to operate fashions and genomics basis fashions on public benchmarks and on the Ntv3 Benchmark, which comprises 106 standardized lengthy vary downstream duties with 32 kb enter and base decision outputs.
- The identical spine helps controllable enhancer design validated with STARR seq: NTv3 might be advantageous tuned as a controllable generative mannequin utilizing masked diffusion language modeling to design enhancer sequences with specified exercise ranges and promoter selectivity, and these designs are validated experimentally with STARR seq assays that verify the meant exercise ordering and improved promoter specificity.
Try the Repo, Mannequin on HF and Technical particulars. Additionally, be at liberty to comply with us on Twitter and don’t neglect to hitch our 100k+ ML SubReddit and Subscribe to our E-newsletter. Wait! are you on telegram? now you may be part of us on telegram as properly.
Asif Razzaq is the CEO of Marktechpost Media Inc.. As a visionary entrepreneur and engineer, Asif is dedicated to harnessing the potential of Synthetic Intelligence for social good. His most up-to-date endeavor is the launch of an Synthetic Intelligence Media Platform, Marktechpost, which stands out for its in-depth protection of machine studying and deep studying information that’s each technically sound and simply comprehensible by a large viewers. The platform boasts of over 2 million month-to-month views, illustrating its recognition amongst audiences.